SMART Guidelines Starter Kit
2.1.0 - ci-build
SMART Guidelines Starter Kit
2.1.0 - ci-build
SMART Guidelines Starter Kit, published by WHO. This guide is not an authorized publication; it is the continuous build for version 2.1.0 built by the FHIR (HL7® FHIR® Standard) CI Build. This version is based on the current content of https://github.com/WorldHealthOrganization/smart-ig-starter-kit/tree/main and changes regularly. See the Directory of published versions
Logical models represent the data structures in the Digital Dictionary. This is a computable representation that is independent of any physical contraints such as specific FHIR Releases or profile versions. The use of FHIR logical models allows metadata to be structured, computable, and interoperable, for the purposes of governance and checking.
Creating a FHIR logical model entails capturing the elements in the Data Dictionary, with their description, terminology and cardinality constraints.
Each Logical Model is defined by the following data:
Logical models relate to other models in 3 aspects:
input/[fsh/]modelsThis is the overview of the activities and related artifacts used:
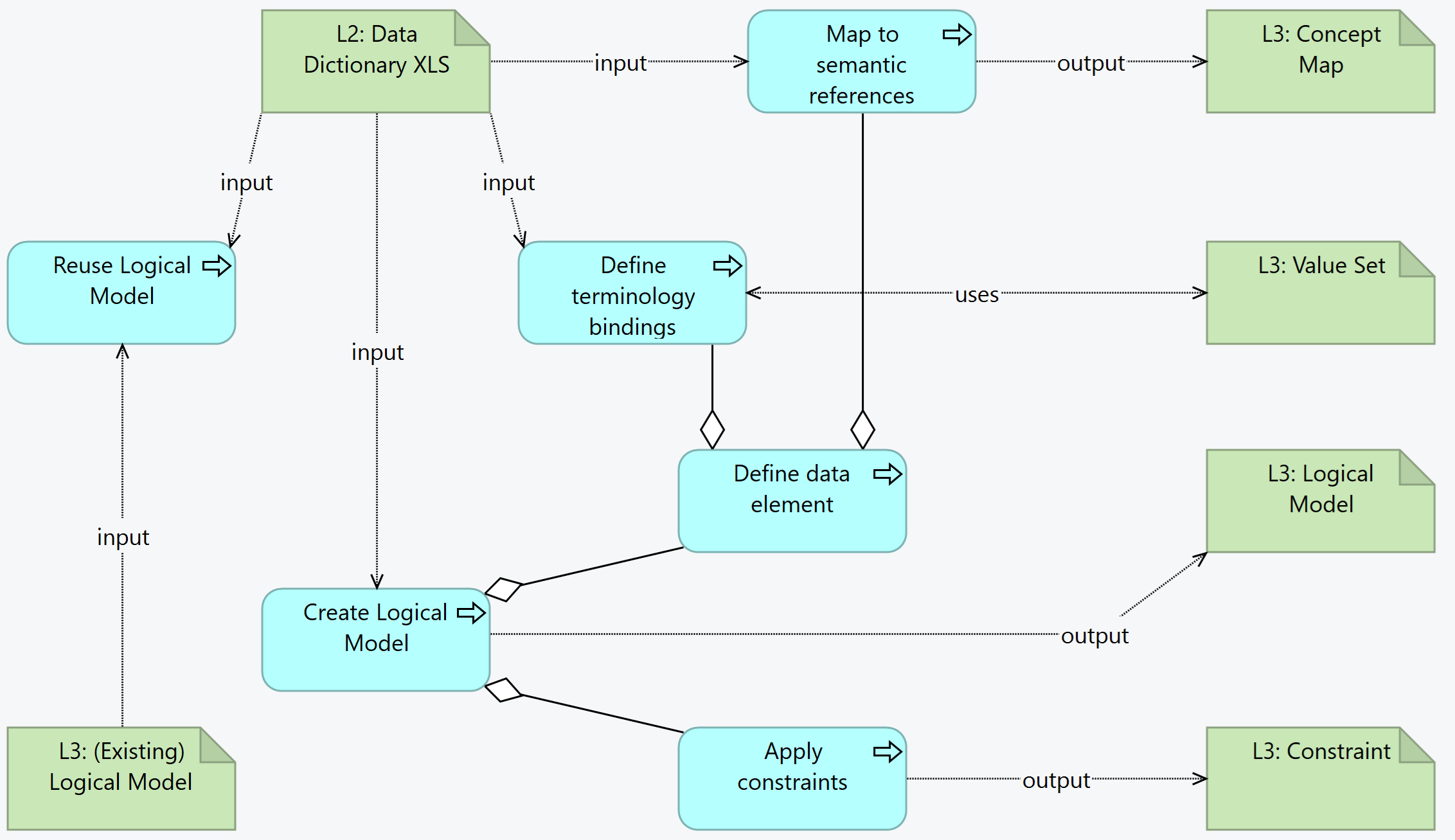
Logical models define data structures. They can be reused, which means that if an existing model is found that is similar to the intended model, it is possible to:
For example, "Client Registration" is a dataset that is similar across different vaccination processes. L3 authors may create a "Client Registration" logical data model with the common elements and create specific models for each antigen with any specific data elements.
Inheritance is preferred for consistency, but the advantage should be checked against the dependency added - if the model that is being authored is expected to increasingly differ from the "parent" model, it may be best to consider a specialization or a new model.
Another possibility is the use of common structures - for example person identification - with given and family names, identifier(s), etc. These can be used as data types or subsets of a logical model.
L3 authors should take into consideration that reused models are version-sensitive and any changes should take this versioning into consideration.
When reusing an existing model, L3 authors are recommended to produce a UML-like diagram or any other illustration that depicts this reuse.
The Data models in the L3 are associated with a data structure for exchange or for a given process. For example "capture vaccination data". L2 authors are expected to follow that rule: one tab is assumed to be one data structure, i.e. a logical data model.
This is the "atomic" unit of exchange or use in the L3. Some factors may influence the split of a logical model into several functional blocks, besides the specialization, reference and usage described above. For example:
The logical model name has the name of the tab.
Creating the logical model from a DAK consists in creating the data structure, linking the elements to the common concept identifiers or, if that is not possible, to the internal unique concept identifiers (e.g. DE1, etc.). Additionally, assigning value sets (creating them when needed), and capturing any constraints that are present in the L2.
To start creating the logical model, an intake validation is useful, although it can be done simultaneously done with the editing of the logical model:
For each line in the tab, there shall be one data element in the logical model (represented here by differential.element[*]):
Every element SHALL have a WHO identifier e.g. TB.DE1.
differential.element[*].short): same as element labelDE1.3 are persisted as a mapping.Description and Definition: differential.element[*].definition
Multiple Choice Type (if applicable): This means the element may have several possible values (as in a multiple-choice question). This corresponds to a * in cardinality.
Data Type is captured in differential.element[*].type
Input Options: is captured in the bindings - a value set for the options
Input Option Groupings: this is a grouper for the rows that have a common group.
Quantity Sub-type (integer, decimal, duration)
differential.element[*].commentRequired is captured in cardinality
Conditionality can be explained as a comment in the data element, or as a formal constraint/invariant.
code[WHO] = IMMZ.DE9For bindings, check whether the value set already exists in an upstream package.
When adding valuesets, the binding strength should be considered:
Semantic mapping is done with ConceptMaps, establishing the relationship between the DAK elements and reference terminologies, central dictionary, or other DAKS.
As with all FHIR Conformance resources, change management is critical. Do not set the version element of LogicalModels defined in the SMART Guideline, the version element will be set by the publication process. See the versioning topic for more information on change management.
| Tool | Usage | Doc |
|---|---|---|
| Sushi | Create LMs with fsh syntax | HL7 Spec Sushi Documentation |
SMART Guidelines - Immunizations (Measles): Logical Models